Vivern – A Virtual Environment for Multiscale Visualization and Modeling of DNA Nanostructures
David Kuťák, Matias Nicolás Selzer, Jan Byška, María Luján Ganuza, Ivan Barišić, Barbora Kozlíková, Haichao Miao
View presentation:2022-10-19T20:57:00ZGMT-0600Change your timezone on the schedule page
2022-10-19T20:57:00Z
Prerecorded Talk
The live footage of the talk, including the Q&A, can be viewed on the session page, DNA/Genome and Molecular Data/Vis.
Fast forward
Keywords
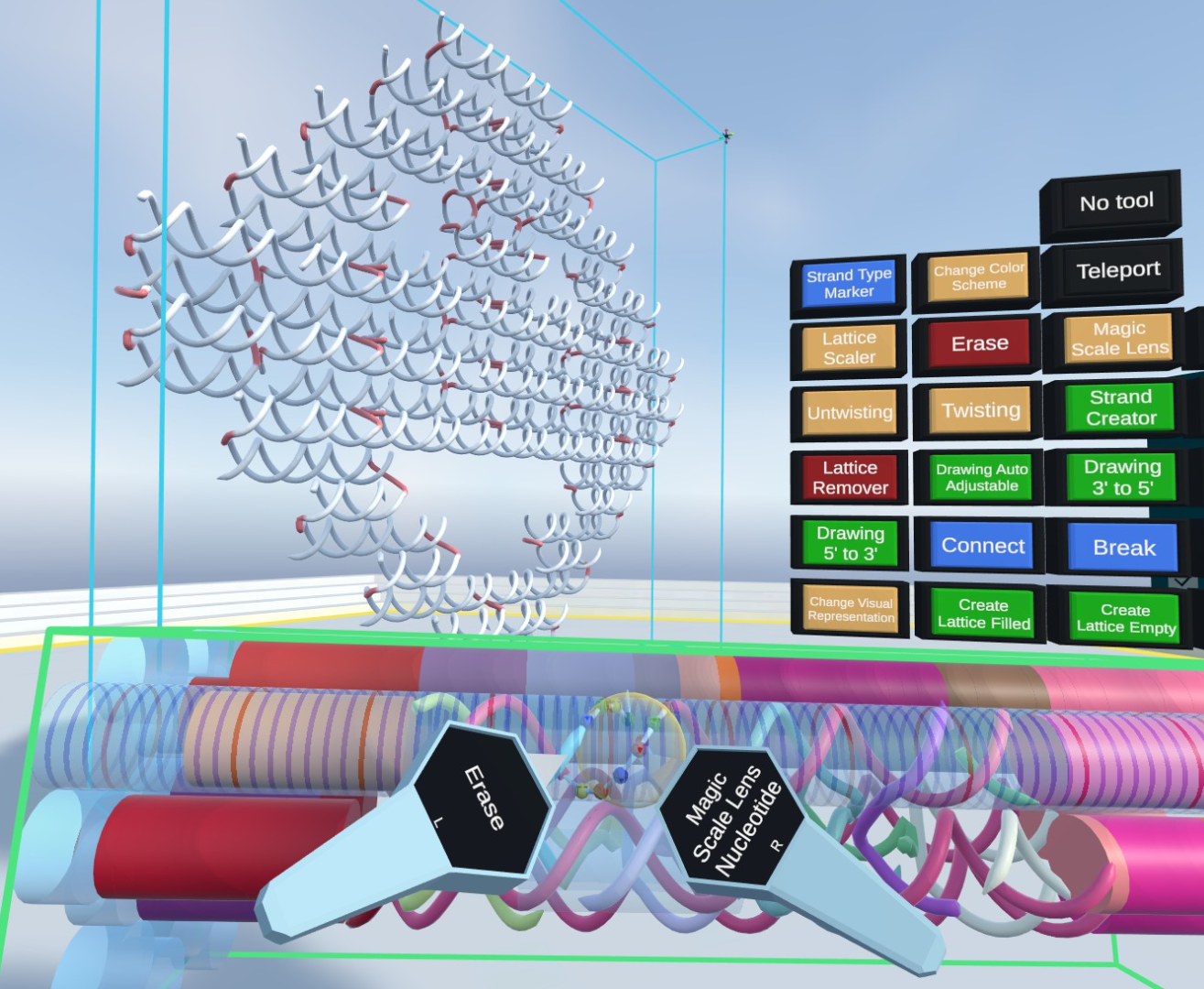
Virtual reality, abstraction, DNA origami, nanostructures, visualization, focus+context, interaction, in silico modeling, nanotechnology, multiscale, magic scale lens
Abstract
DNA nanostructures offer promising applications, particularly in the biomedical domain, as they can be used for targeted drug delivery, construction of nanorobots, or as a basis for molecular motors. One of the most prominent techniques for assembling these structures is DNA origami. Nowadays, desktop applications are used for the in silico design of such structures. However, as such structures are often spatially complex, their assembly and analysis are complicated. Since virtual reality was proven to be advantageous for such spatial-related tasks and there are no existing VR solutions focused on this domain, we propose Vivern, a VR application that allows domain experts to design and visually examine DNA origami nanostructures. Our approach presents different abstracted visual representations of the nanostructures, various color schemes, and an ability to place several DNA nanostructures and proteins in one environment, thus allowing for the detailed analysis of complex assemblies. We also present two novel examination tools, the Magic Scale Lens and the DNA Untwister, that allow the experts to visually embed different representations into local regions to preserve the context and support detailed investigation. To showcase the capabilities of our solution, prototypes of novel nanodevices conceptualized by our collaborating experts, such as DNA-protein hybrid structures and DNA origami superstructures, are presented. Finally, the results of two rounds of evaluations are summarized. They demonstrate the advantages of our solution, especially for scenarios where current desktop tools are very limited, while also presenting possible future research directions.